pyNastran/op2¶
This is the pyNastran.op2.rst file.
op2 Module¶
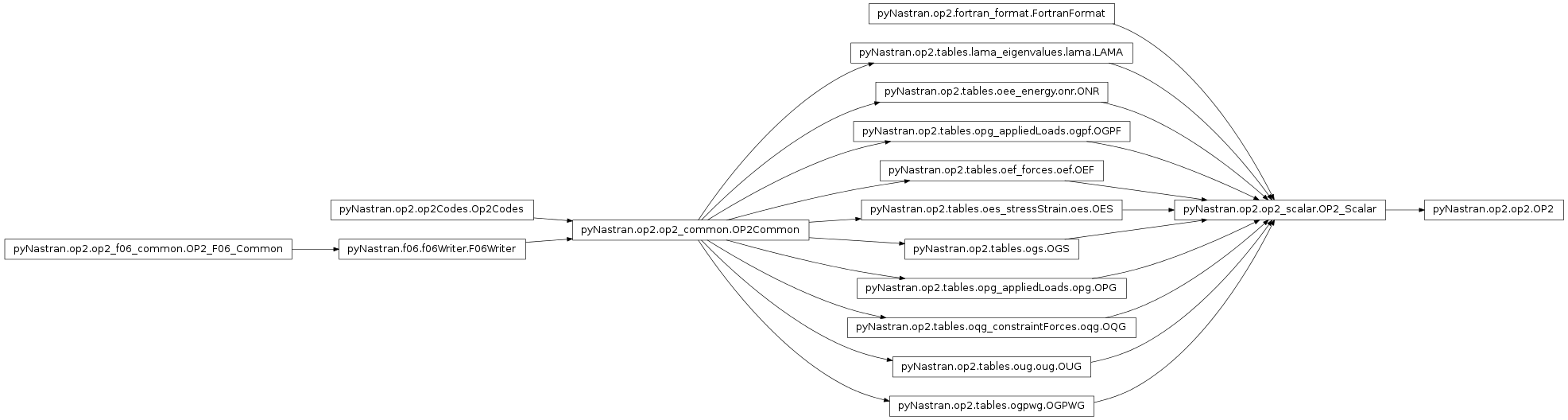
Main OP2 class
-
class
pyNastran.op2.op2.OP2(debug=True, log=None, debug_file=None)[source]¶ Bases:
pyNastran.op2.op2_scalar.OP2_ScalarInitializes the OP2 object
Parameters: - debug – enables the debug log and sets the debug in the logger (default=False)
- log – a logging object to write debug messages to (.. seealso:: import logging)
- debug_file – sets the filename that will be written to (default=None -> no debug)
Methods
-
combine_results(combine=True)[source]¶ we want the data to be in the same format and grouped by subcase, so we take
stress = { (1, 'SUPERELEMENT 0') : result1, (1, 'SUPERELEMENT 10') : result2, (1, 'SUPERELEMENT 20') : result3, (2, 'SUPERELEMENT 0') : result4, }
and convert it to:
stress = { 1 : result1 + result2 + results3, 2 : result4, }
-
read_op2(op2_filename=None, vectorized=True, combine=True)[source]¶ Starts the OP2 file reading :param op2_filename: the op2_filename (default=None -> popup) :param vectorized: should the vectorized objects be used (default=True) :param combine: should objects be isubcase based (True) or
(isubcase, subtitle) based (False) The second will be used for superelements regardless of the option (default=True)
-
set_as_vectorized(ask=False)[source]¶ Enables vectorization
The code will degenerate to dictionary based results when a result does not support vectorization.
Vectorization is always True here. :param ask: Do you want to see a GUI of result types.
Case # Vectorization Ask Read Modes 1 True True 1, 2 2 True False 1, 2 3 False True 1, 2 4 False False 0
op2_scalar Module¶
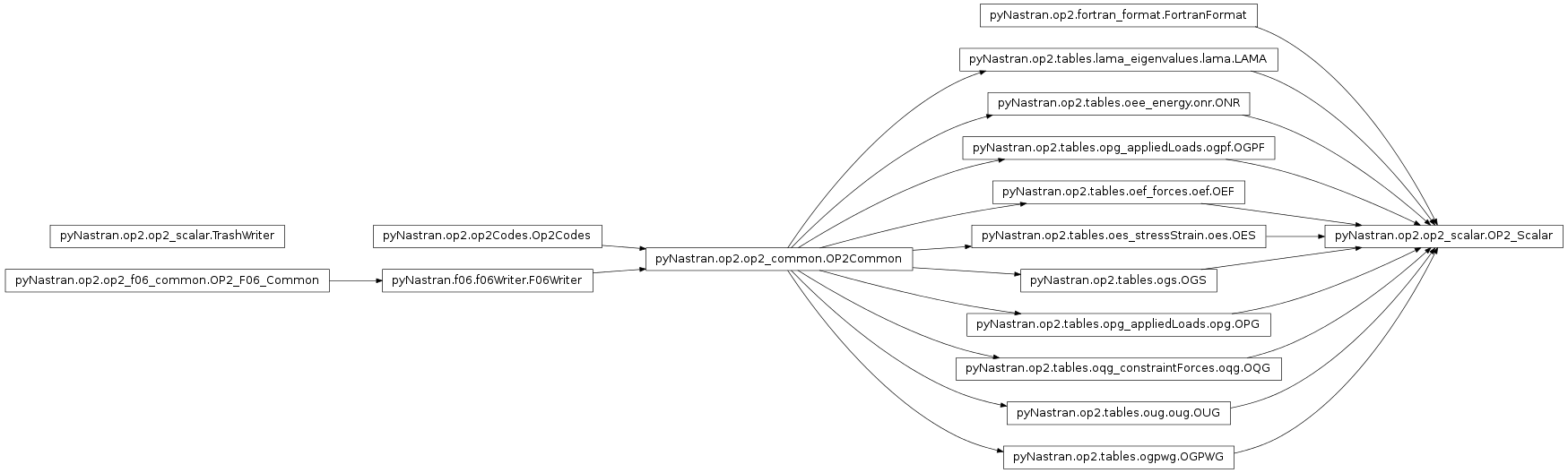
Defines the OP2 class.
-
class
pyNastran.op2.op2_scalar.OP2_Scalar(debug=False, log=None, debug_file=None)[source]¶ Bases:
pyNastran.op2.tables.lama_eigenvalues.lama.LAMA,pyNastran.op2.tables.oee_energy.onr.ONR,pyNastran.op2.tables.opg_appliedLoads.ogpf.OGPF,pyNastran.op2.tables.oef_forces.oef.OEF,pyNastran.op2.tables.oes_stressStrain.oes.OES,pyNastran.op2.tables.ogs.OGS,pyNastran.op2.tables.opg_appliedLoads.opg.OPG,pyNastran.op2.tables.oqg_constraintForces.oqg.OQG,pyNastran.op2.tables.oug.oug.OUG,pyNastran.op2.tables.ogpwg.OGPWG,pyNastran.op2.fortran_format.FortranFormatDefines an interface for the Nastran OP2 file.
Methods
Initializes the OP2_Scalar object
Parameters: - debug – enables the debug log and sets the debug in the logger (default=False)
- log – a logging object to write debug messages to (.. seealso:: import logging)
- debug_file – sets the filename that will be written to (default=None -> no debug)
Methods
-
finish()[source]¶ Clears out the data members contained within the self.words variable. This prevents mixups when working on the next table, but otherwise has no effect.
-
read_op2(op2_filename=None)[source]¶ Starts the OP2 file reading
Parameters: op2_filename – the op2 file op2_filename Description None a dialog is popped up string the path is used
-
read_table_name(rewind=False, stop_on_failure=True)[source]¶ Reads the next OP2 table name (e.g. OUG1, OES1X1)
fortran_format Module¶

-
class
pyNastran.op2.fortran_format.FortranFormat[source]¶ Bases:
objectParameters: self – the OP2 object pointer Methods
-
get_nmarkers(n, rewind=True)[source]¶ Gets n markers, so if n=2, it will get 2 markers.
Parameters: - self – the OP2 object pointer
- n – number of markers to get
- rewind – should the file be returned to the starting point
Retval markers: list of [1, 2, 3, ...] markers
-
goto(n)[source]¶ Jumps to position n in the file
Parameters: - self – the OP2 object pointer
- n – the position to goto
-
isAllSubcases= None¶ stores if the user entered [] for iSubcases
-
is_valid_subcase()[source]¶ Lets the code check whether or not to read a subcase
Parameters: self – the OP2 object pointer Retval is_valid: should this subcase defined by self.isubcase be read?
-
passer(data)[source]¶ dummy function used for unsupported tables :param self: the OP2 object pointer
-
read_block()[source]¶ - Reads a block following a pattern of:
- [nbytes, data, nbytes]
Retval data: the data in binary
-
read_markers(markers)[source]¶ Gets specified markers, where a marker has the form of [4, value, 4]. The “marker” corresponds to the value, so 3 markers takes up 9 integers. These are used to indicate position in the file as well as the number of bytes to read.
Parameters: - self – the OP2 object pointer
- markers – markers to get; markers = [-10, 1]
-
skip_block()[source]¶ - Skips a block following a pattern of:
- [nbytes, data, nbytes]
Parameters: self – the OP2 object pointer Retval data: since data can never be None, a None value indicates something bad happened.
-
op2_codes Module¶
op2_helper Module¶
-
pyNastran.op2.op2_helper.polar_to_real_imag(mag, phase)[source]¶ Converts magnitude-phase to real-imaginary so all complex results are consistent
Parameters: - mag – magnitude c^2
- phase – phase angle phi (degrees; theta)
Returns realValue: the real component a of a+bi
Returns imagValue: the imaginary component b of a+bi
op2_common Module¶

-
class
pyNastran.op2.op2_common.OP2Common[source]¶ Bases:
pyNastran.op2.op2Codes.Op2Codes,pyNastran.f06.f06Writer.F06WriterMethods
-
ID= None¶ the corresponding piece to isubcase used only for SORT2 (not supported)
-
add_data_parameter(data, var_name, Type, field_num, applyNonlinearFactor=True, fixDeviceCode=False, add_to_dict=True)[source]¶
-
binary_debug= None¶ op2 debug file or None (for self.debug=False)
-
create_transient_object(storageObj, classObj, is_cid=False, debug=False)[source]¶ Creates a transient object (or None if the subcase should be skippied).
Parameters: - storageName – the name of the dictionary to store the object in (e.g. ‘displacements’)
- classObj – the class object to instantiate
- debug – developer debug
Note
dt can also be load_step depending on the class
-
data_code= None¶ the storage dictionary that is passed to OP2 objects (e.g. DisplacementObject) the key-value pairs are extracted and used to generate dynamic self variables for the OP2 objects
-
debug= None¶ should the op2 debugging file be written
-
expected_times= None¶ the list/set/tuple of times/modes/frequencies that should be read currently unused
-
is_vectorized= None¶ bool
-
isubcase= None¶ current subcase ID non-transient (SOL101) cases have isubcase set to None transient (or frequency/modal) cases have isubcase set to a int/float value
-
read_mode= None¶ flag for vectorization 0 - no vectorization 1 - first pass 2 - second pass
-
result_names= None¶ the results
-
subcases= None¶ set of all the subcases that have been found
-
table_name= None¶ The current table_name (e.g. OES1) None indicates no table_name has been read
-
words= None¶ the list of “words” on a subtable 3
-
op2_f06_common Module¶

data_in_material_coord Module¶
vector_utils Module¶
-
pyNastran.op2.vector_utils.abs_max_min_global(values)[source]¶ This is useful for figuring out absolute max or min principal stresses across single/multiple elements and finding a global max/min value.
Parameters: values (common NDARRAY/list/tuple shapes: 1. [nprincipal_stresses] 2. [nelements, nprincipal_stresses]) – an ND-array of values Returns abs_max_mins: an array of the max or min principal stress - nvalues >= 1
>>> element1 = [0.0, -1.0, 2.0] # 2.0 >>> element2 = [0.0, -3.0, 2.0] # -3.0 >>> values = abs_max_min_global([element1, element2]) >>> values -3.0
>>> element1 = [0.0, -1.0, 2.0] # 2.0 >>> values = abs_max_min_global([element1]) >>> values 2.0
Note
[3.0, 2.0, -3.0] will return 3.0, and [-3.0, 2.0, 3.0] will return 3.0
-
pyNastran.op2.vector_utils.abs_max_min_vector(values)[source]¶ This is useful for figuring out principal stresses across multiple elements.
Parameters: values (NDARRAY shape=[nelements, nprincipal_stresses]) – an array of values, where the rows are interated over and the columns are going to be compressed Returns abs_max_mins: an array of the max or min principal stress - ::
>>> element1 = [0.0, 1.0, 2.0] # 2.0 >>> element2 = [0.0, -1.0, 2.0] # 2.0 >>> element3 = [0.0, -3.0, 2.0] # -3.0 >>> values = [element1 element2, element3] >>> values0 = abs_max_min_vectorized(values) >>> values0 [2.0, 2.0, -3.0]
Note
[3.0, 2.0, -3.0] will return 3.0, and [-3.0, 2.0, 3.0] will return 3.0
- resultObjects Package
- tables Package
grid_point_weightModuleogsModuleogpwgModule
- test Package