op2 Package¶
This is the pyNastran.op2.rst file.
op2 Module¶
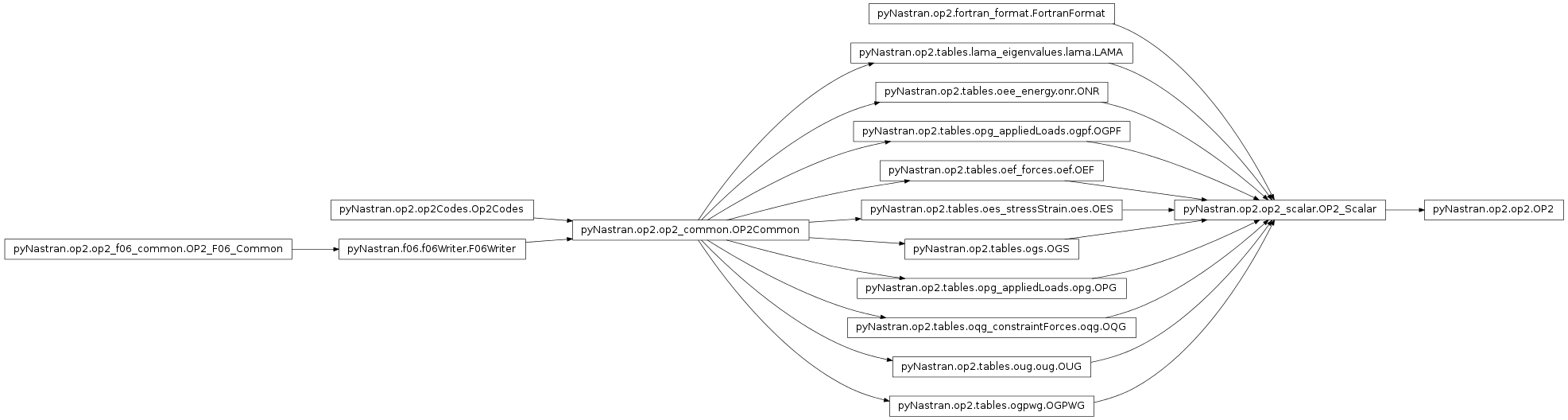
Main OP2 class
- class pyNastran.op2.op2.OP2(debug=True, log=None, debug_file=None)[source]¶
Bases: pyNastran.op2.op2_scalar.OP2_Scalar
Initializes the OP2 object
Parameters: - debug – enables the debug log and sets the debug in the logger (default=False)
- log – a logging object to write debug messages to (.. seealso:: import logging)
- debug_file – sets the filename that will be written to (default=None -> no debug)
- combine_results(combine=True)[source]¶
we want the data to be in the same format and grouped by subcase, so we take
stress = { (1, 'SUPERELEMENT 0') : result1, (1, 'SUPERELEMENT 10') : result2, (1, 'SUPERELEMENT 20') : result3, (2, 'SUPERELEMENT 0') : result4, }
and convert it to:
stress = { 1 : result1 + result2 + results3, 2 : result4, }
- read_op2(op2_filename=None, vectorized=True, combine=True)[source]¶
Starts the OP2 file reading :param op2_filename: the op2_filename (default=None -> popup) :param vectorized: should the vectorized objects be used (default=True) :param combine: should objects be isubcase based (True) or
(isubcase, subtitle) based (False) The second will be used for superelements regardless of the option (default=True)
- set_as_vectorized(ask=False)[source]¶
Enables vectorization
The code will degenerate to dictionary based results when a result does not support vectorization.
Vectorization is always True here. :param ask: Do you want to see a GUI of result types.
Case # Vectorization Ask Read Modes 1 True True 1, 2 2 True False 1, 2 3 False True 1, 2 4 False False 0 - Vectorization - A storage structure that allows for faster read/access
speeds and better memory usage, but comes with a more difficult to use data structure.
It limits the node IDs to all be integers (e.g. element centroid). Composite plate elements (even for just CTRIA3s) with an inconsistent number of layers will have a more difficult data structure. Models with solid elements of mixed type will also be more complicated (or potentially split up).
- Scanning - a quick check used to figure out how many results to process
- that takes almost no time
Reading - process the op2 data Build - call the __init__ on a results object (e.g. DisplacementObject) Start Over - Go to the start of the op2 file Ask - launch a GUI dialog to let the user click which results to load
- The default OP2 dictionary based-approach with no asking GUI
- The first read of a result to get the shape of the data (and result types if vectorization=True or result types if vectorization=False)
- The second read of a result to get the results
- Scan the block to get the size, build the object (read_mode=1), ask the user, start over, fill the objects (read_mode=2). Degenerate to read_mode=0 when read_mode=2 cannot be used based upon the value of ask.
- Same as case #1, but don’t ask the user. Scan the block to get the size, build the object (read_mode=1), start over, fill the objects (read_mode=2).
- Scan the block to get the object types (read_mode=1), ask the user, build the object & fill it (read_mode=2)
- Read the block to get the size, build the object & fill it (read_mode=0)
op2_scalar Module¶
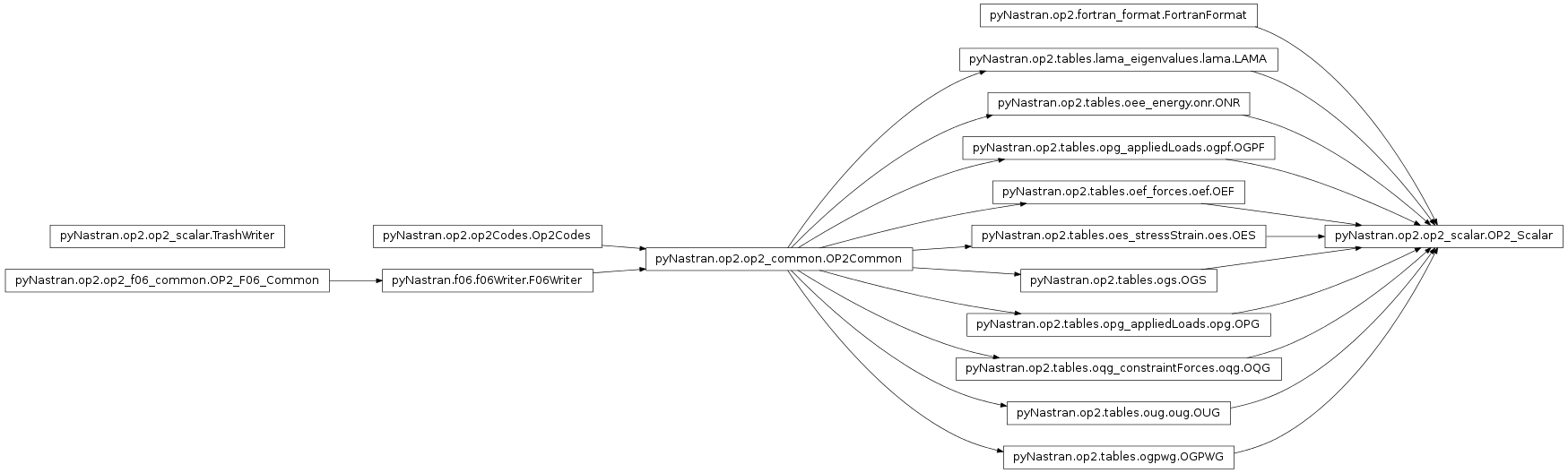
Defines the OP2 class.
- class pyNastran.op2.op2_scalar.OP2_Scalar(debug=False, log=None, debug_file=None)[source]¶
Bases: pyNastran.op2.tables.lama_eigenvalues.lama.LAMA, pyNastran.op2.tables.oee_energy.onr.ONR, pyNastran.op2.tables.opg_appliedLoads.ogpf.OGPF, pyNastran.op2.tables.oef_forces.oef.OEF, pyNastran.op2.tables.oes_stressStrain.oes.OES, pyNastran.op2.tables.ogs.OGS, pyNastran.op2.tables.opg_appliedLoads.opg.OPG, pyNastran.op2.tables.oqg_constraintForces.oqg.OQG, pyNastran.op2.tables.oug.oug.OUG, pyNastran.op2.tables.ogpwg.OGPWG, pyNastran.op2.fortran_format.FortranFormat
Defines an interface for the Nastran OP2 file.
Initializes the OP2_Scalar object
Parameters: - debug – enables the debug log and sets the debug in the logger (default=False)
- log – a logging object to write debug messages to (.. seealso:: import logging)
- debug_file – sets the filename that will be written to (default=None -> no debug)
- _print_month(month, day, year, zero, one)[source]¶
Creates the self.date attribute from the 2-digit year.
Parameters: - month – the month (integer <= 12)
- day – the day (integer <= 31)
- year – the day (integer <= 99)
- zero – a dummy integer (???)
- one – a dummy integer (???)
- _read_dit()[source]¶
Reads the DIT table (poorly). The DIT table stores information about table cards (e.g. TABLED1, TABLEM1).
- _read_kelm()[source]¶
Todo
this table follows a totally different pattern...
The KELM table stores information about the K matrix???
- _read_pcompts()[source]¶
Reads the PCOMPTS table (poorly). The PCOMPTS table stores information about the PCOMP cards???
- _read_tables(table_name)[source]¶
Reads all the geometry/result tables. The OP2 header is not read by this function.
Parameters: table_name – the first table’s name
- _skip_pcompts()[source]¶
Reads the PCOMPTS table (poorly). The PCOMPTS table stores information about the PCOMP cards???
- _skip_table_helper()[source]¶
Skips the majority of geometry/result tables as they follow a very standard format. Other tables don’t follow this format.
- finish()[source]¶
Clears out the data members contained within the self.words variable. This prevents mixups when working on the next table, but otherwise has no effect.
- read_op2(op2_filename=None)[source]¶
Starts the OP2 file reading
Parameters: op2_filename – the op2 file op2_filename Description None a dialog is popped up string the path is used
- read_table_name(rewind=False, stop_on_failure=True)[source]¶
Reads the next OP2 table name (e.g. OUG1, OES1X1)
fortran_format Module¶

- class pyNastran.op2.fortran_format.FortranFormat[source]¶
Bases: object
Parameters: self – the OP2 object pointer - _get_record_length()[source]¶
The record length helps us figure out data block size, which is used to quickly size the arrays. We just need a bit of meta data and can jump around quickly.
- _read_subtable_results(table4_parser, record_len)[source]¶
# if reading the data # 0 - non-vectorized # 1 - 1st pass to size the array (vectorized) # 2 - 2nd pass to read the data (vectorized)
- _stream_record(debug=True)[source]¶
Creates a “for” loop that keeps giving us records until we’re done.
Parameters: self – the OP2 object pointer
- get_nmarkers(n, rewind=True)[source]¶
Gets n markers, so if n=2, it will get 2 markers.
Parameters: - self – the OP2 object pointer
- n – number of markers to get
- rewind – should the file be returned to the starting point
Retval markers: list of [1, 2, 3, ...] markers
- goto(n)[source]¶
Jumps to position n in the file
Parameters: - self – the OP2 object pointer
- n – the position to goto
- isAllSubcases = None¶
stores if the user entered [] for iSubcases
- is_valid_subcase()[source]¶
Lets the code check whether or not to read a subcase
Parameters: self – the OP2 object pointer Retval is_valid: should this subcase defined by self.isubcase be read?
- passer(data)[source]¶
dummy function used for unsupported tables :param self: the OP2 object pointer
- read_block()[source]¶
- Reads a block following a pattern of:
- [nbytes, data, nbytes]
Retval data: the data in binary
- read_markers(markers)[source]¶
Gets specified markers, where a marker has the form of [4, value, 4]. The “marker” corresponds to the value, so 3 markers takes up 9 integers. These are used to indicate position in the file as well as the number of bytes to read.
Parameters: - self – the OP2 object pointer
- markers – markers to get; markers = [-10, 1]
- skip_block()[source]¶
- Skips a block following a pattern of:
- [nbytes, data, nbytes]
Parameters: self – the OP2 object pointer Retval data: since data can never be None, a None value indicates something bad happened.
op2_helper Module¶
- pyNastran.op2.op2_helper.polar_to_real_imag(mag, phase)[source]¶
Converts magnitude-phase to real-imaginary so all complex results are consistent
Parameters: - mag – magnitude c^2
- phase – phase angle phi (degrees; theta)
Returns realValue: the real component a of a+bi
Returns imagValue: the imaginary component b of a+bi
op2_common Module¶

- class pyNastran.op2.op2_common.OP2Common[source]¶
Bases: pyNastran.op2.op2Codes.Op2Codes, pyNastran.f06.f06Writer.F06Writer
- ID = None¶
the corresponding piece to isubcase used only for SORT2 (not supported)
- _parse_sort_code()[source]¶
sort_code sort_bits 0 [0, 0, 0] 1 [0, 0, 1] 2 [0, 1, 0] 3 [0, 1, 1] ... ... 7 [1, 1, 1] - ::
sort_code = 0 -> sort_bits = [0,0,0] sort_code = 1 -> sort_bits = [0,0,1] sort_code = 2 -> sort_bits = [0,1,0] sort_code = 3 -> sort_bits = [0,1,1] etc. sort_code = 7 -> sort_bits = [1,1,1]
sort_bits[0] = 0 -> is_sort1=True isSort2=False sort_bits[1] = 0 -> isReal=True isReal/Imaginary=False sort_bits[2] = 0 -> isSorted=True isRandom=False
- _read_table(data, result_name, storage_obj, real_obj, complex_obj, real_vector, complex_vector, node_elem, random_code=None, is_cid=False)[source]¶
- _table_specs()[source]¶
Value Sort Type Data Format Random ? 0 SORT1 Real No 1 SORT1 Complex No 2 SORT2 Real No 3 SORT2 Complex No 4 SORT1 Real Yes 5 SORT2 Real Yes
- add_data_parameter(data, var_name, Type, field_num, applyNonlinearFactor=True, fixDeviceCode=False, add_to_dict=True)[source]¶
- binary_debug = None¶
op2 debug file or None (for self.debug=False)
- create_transient_object(storageObj, classObj, is_cid=False, debug=False)[source]¶
Creates a transient object (or None if the subcase should be skippied).
Parameters: - storageName – the name of the dictionary to store the object in (e.g. ‘displacements’)
- classObj – the class object to instantiate
- debug – developer debug
Note
dt can also be load_step depending on the class
- data_code = None¶
the storage dictionary that is passed to OP2 objects (e.g. DisplacementObject) the key-value pairs are extracted and used to generate dynamic self variables for the OP2 objects
- debug = None¶
should the op2 debugging file be written
- expected_times = None¶
the list/set/tuple of times/modes/frequencies that should be read currently unused
- is_vectorized = None¶
bool
- isubcase = None¶
current subcase ID non-transient (SOL101) cases have isubcase set to None transient (or frequency/modal) cases have isubcase set to a int/float value
- read_mode = None¶
flag for vectorization 0 - no vectorization 1 - first pass 2 - second pass
- result_names = None¶
the results
- subcases = None¶
set of all the subcases that have been found
- table_name = None¶
The current table_name (e.g. OES1) None indicates no table_name has been read
- words = None¶
the list of “words” on a subtable 3
op2_f06_common Module¶

vector_utils Module¶
- pyNastran.op2.vector_utils.abs_max_min_global(values)[source]¶
This is useful for figuring out absolute max or min principal stresses across single/multiple elements and finding a global max/min value.
Parameters: values (common NDARRAY/list/tuple shapes: 1. [nprincipal_stresses] 2. [nelements, nprincipal_stresses]) – an ND-array of values Returns abs_max_mins: an array of the max or min principal stress - nvalues >= 1
>>> element1 = [0.0, -1.0, 2.0] # 2.0 >>> element2 = [0.0, -3.0, 2.0] # -3.0 >>> values = abs_max_min_global([element1, element2]) >>> values -3.0
>>> element1 = [0.0, -1.0, 2.0] # 2.0 >>> values = abs_max_min_global([element1]) >>> values 2.0
Note
[3.0, 2.0, -3.0] will return 3.0, and [-3.0, 2.0, 3.0] will return 3.0
- pyNastran.op2.vector_utils.abs_max_min_vector(values)[source]¶
This is useful for figuring out principal stresses across multiple elements.
Parameters: values (NDARRAY shape=[nelements, nprincipal_stresses]) – an array of values, where the rows are interated over and the columns are going to be compressed Returns abs_max_mins: an array of the max or min principal stress - ::
>>> element1 = [0.0, 1.0, 2.0] # 2.0 >>> element2 = [0.0, -1.0, 2.0] # 2.0 >>> element3 = [0.0, -3.0, 2.0] # -3.0 >>> values = [element1 element2, element3] >>> values0 = abs_max_min_vectorized(values) >>> values0 [2.0, 2.0, -3.0]
Note
[3.0, 2.0, -3.0] will return 3.0, and [-3.0, 2.0, 3.0] will return 3.0
- resultObjects Package
- tables Package
- test Package
